Singapore has experienced substantial habitat and biodiversity loss since 1819. Nevertheless, the last century has also seen some forest recovery, as well as the natural re-establishment of several previously extirpated species like oriental pied hornbills and smooth-coated otters. Animals that can fly and swim can re-establish from source populations in Malaysia, but snakes and lizards—also known as squamates—are not able to recolonise as easily. How can we use mathematical models to think about which species to reintroduce to recovering forests? In 2023, we published a paper applying the novel MODGEE (matrix-of-detections-gives-extinction-estimates) model to detailed historical bird, butterfly and plant datasets from Singapore to study extirpation rates for these groups. In a new study, led by Sankar and just published in Biological Conservation, we applied this model to squamates in Singapore.
We compiled a detailed historical dataset for squamates in Singapore (largely based on records from Figueroa et al., 2023) and used the MODGEE model first to generate an extirpation timeline. Our estimated extirpation rate for squamates was 17%, which was lower than previously estimated rates for birds (40%) and butterflies (46%). This implies that squamates may be more resilient to habitat degradation than birds, perhaps because birds generally have lower population densities, and butterflies, perhaps because of butterflies’ reliance on host plants.
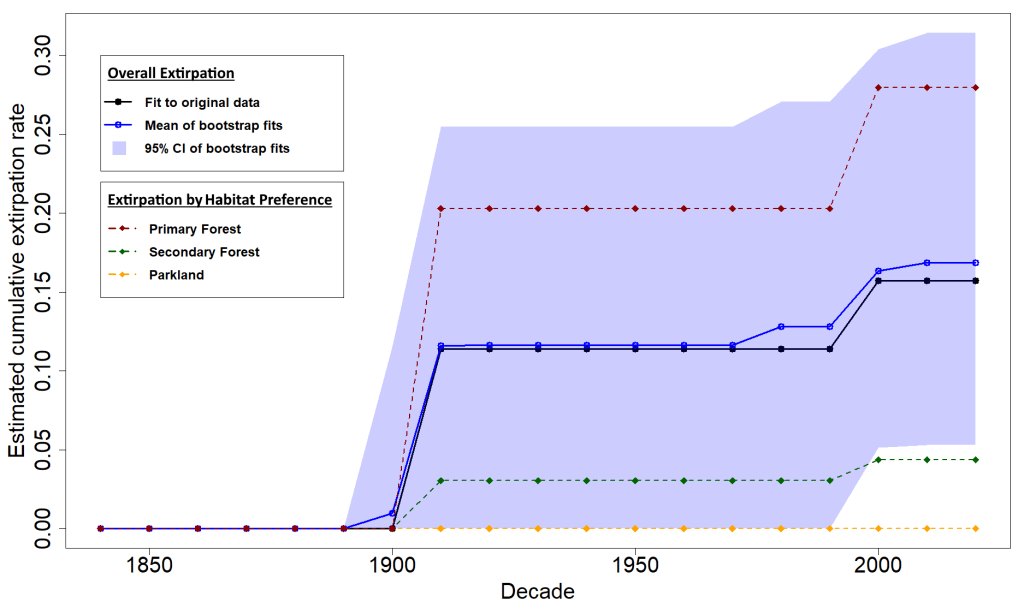
We also used the results to inform a conservation translocation triage for extirpated squamates in Singapore. Given that squamates were never systematically hunted at scale (no pun intended) in Singapore, their extirpation timeline can serve as an indicator of each species’ sensitivity to habitat loss. Since highly sensitive species would have been lost early on in Singapore’s developmental history, recently extirpated species would likely be better candidates for reintroduction to Singapore’s recovering forests. Through this method, we identified the hulk forest gecko (Gekko hulk) as a fitting potential candidate species for reintroduction to Singapore from stock populations in Peninsular Malaysia.






